Empirical Test Statistics¶
In this notebook we will compute test statistics empirically from pseudo-experiment and establish that they behave as assumed in the asymptotic approximation.
[1]:
import numpy as np
import matplotlib.pyplot as plt
import pyhf
np.random.seed(0)
plt.rcParams.update({"font.size": 14})
First, we define the statistical model we will study.
the signal expected event rate is 10 events
the background expected event rate is 100 events
a 10% uncertainty is assigned to the background
The expected event rates are chosen to lie comfortably in the asymptotic regime.
[2]:
model = pyhf.simplemodels.hepdata_like(
signal_data=[10.0], bkg_data=[100.0], bkg_uncerts=[10.0]
)
The test statistics based on the profile likelihood described in arXiv:1007.1727 cover scanarios for both
POI allowed to float to negative values (unbounded; \(\mu \in [-10, 10]\))
POI constrained to non-negative values (bounded; \(\mu \in [0,10]\))
For consistency, test statistics (\(t_\mu, q_\mu\)) associated with bounded POIs are usually denoted with a tilde (\(\tilde{t}_\mu, \tilde{q}_\mu\)).
We set up the bounds for the fit as follows
[3]:
unbounded_bounds = model.config.suggested_bounds()
unbounded_bounds[model.config.poi_index] = (-10, 10)
bounded_bounds = model.config.suggested_bounds()
Next we draw some synthetic datasets (also referrerd to as “toys” or pseudo-experiments). We will “throw” 300 toys:
[4]:
true_poi = 1.0
n_toys = 300
toys = model.make_pdf(pyhf.tensorlib.astensor([true_poi, 1.0])).sample((n_toys,))
In the asymptotic treatment the test statistics are described as a function of the data’s best-fit POI value \(\hat\mu\).
So let’s run some fits so we can plots the empirical test statistics against \(\hat\mu\) to observed the emergence of the asymptotic behavior.
[5]:
pars = np.asarray(
[pyhf.infer.mle.fit(toy, model, par_bounds=unbounded_bounds) for toy in toys]
)
fixed_params = model.config.suggested_fixed()
We can now calculate all four test statistics described in arXiv:1007.1727
[6]:
test_poi = 1.0
tmu = np.asarray(
[
pyhf.infer.test_statistics.tmu(
test_poi,
toy,
model,
init_pars=model.config.suggested_init(),
par_bounds=unbounded_bounds,
fixed_params=fixed_params,
)
for toy in toys
]
)
[7]:
tmu_tilde = np.asarray(
[
pyhf.infer.test_statistics.tmu_tilde(
test_poi,
toy,
model,
init_pars=model.config.suggested_init(),
par_bounds=bounded_bounds,
fixed_params=fixed_params,
)
for toy in toys
]
)
[8]:
qmu = np.asarray(
[
pyhf.infer.test_statistics.qmu(
test_poi,
toy,
model,
init_pars=model.config.suggested_init(),
par_bounds=unbounded_bounds,
fixed_params=fixed_params,
)
for toy in toys
]
)
[9]:
qmu_tilde = np.asarray(
[
pyhf.infer.test_statistics.qmu_tilde(
test_poi,
toy,
model,
init_pars=model.config.suggested_init(),
par_bounds=bounded_bounds,
fixed_params=fixed_params,
)
for toy in toys
]
)
Let’s plot all the test statistics we have computed
[10]:
muhat = pars[:, model.config.poi_index]
muhat_sigma = np.std(muhat)
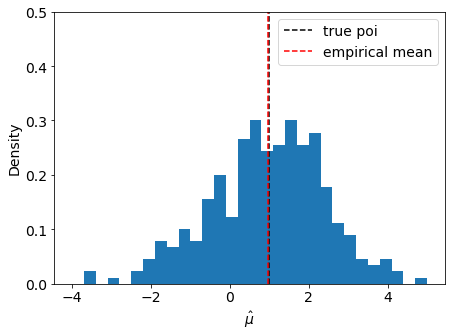
We can check the asymptotic assumption that \(\hat{\mu}\) is distributed normally around it’s true value \(\mu' = 1\)
[11]:
fig, ax = plt.subplots()
fig.set_size_inches(7, 5)
ax.set_xlabel(r"$\hat{\mu}$")
ax.set_ylabel("Density")
ax.set_ylim(top=0.5)
ax.hist(muhat, bins=np.linspace(-4, 5, 31), density=True)
ax.axvline(true_poi, label="true poi", color="black", linestyle="dashed")
ax.axvline(np.mean(muhat), label="empirical mean", color="red", linestyle="dashed")
ax.legend();
Here we define the asymptotic profile likelihood test statistics:
[12]:
def tmu_asymp(mutest, muhat, sigma):
return (mutest - muhat) ** 2 / sigma ** 2
def tmu_tilde_asymp(mutest, muhat, sigma):
a = tmu_asymp(mutest, muhat, sigma)
b = tmu_asymp(mutest, muhat, sigma) - tmu_asymp(0.0, muhat, sigma)
return np.where(muhat > 0, a, b)
def qmu_asymp(mutest, muhat, sigma):
return np.where(
muhat < mutest, tmu_asymp(mutest, muhat, sigma), np.zeros_like(muhat)
)
def qmu_tilde_asymp(mutest, muhat, sigma):
return np.where(
muhat < mutest, tmu_tilde_asymp(mutest, muhat, sigma), np.zeros_like(muhat)
)
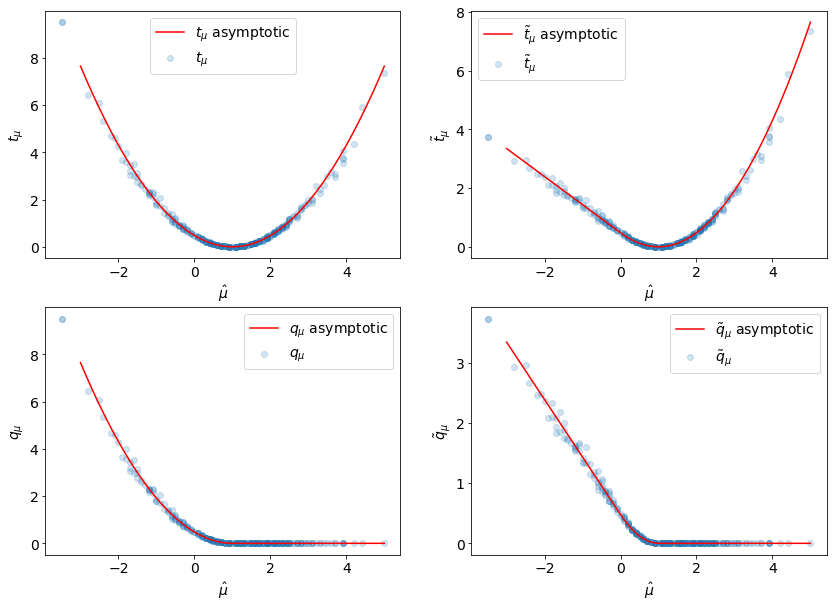
And now we can compare them to the empirical values:
[13]:
muhat_asymp = np.linspace(-3, 5)
fig, axarr = plt.subplots(2, 2)
fig.set_size_inches(14, 10)
labels = [r"$t_{\mu}$", "$\\tilde{t}_{\\mu}$", r"$q_{\mu}$", "$\\tilde{q}_{\\mu}$"]
data = [
(tmu, tmu_asymp),
(tmu_tilde, tmu_tilde_asymp),
(qmu, qmu_asymp),
(qmu_tilde, qmu_tilde_asymp),
]
for ax, (label, d) in zip(axarr.ravel(), zip(labels, data)):
empirical, asymp_func = d
ax.scatter(muhat, empirical, alpha=0.2, label=label)
ax.plot(
muhat_asymp,
asymp_func(1.0, muhat_asymp, muhat_sigma),
label=f"{label} asymptotic",
c="r",
)
ax.set_xlabel(r"$\hat{\mu}$")
ax.set_ylabel(f"{label}")
ax.legend(loc="best")