Multibin Coupled HistoSys¶
[1]:
%pylab inline
Populating the interactive namespace from numpy and matplotlib
[2]:
import logging
import json
import pyhf
from pyhf import Model
logging.basicConfig(level = logging.INFO)
[3]:
def prep_data(sourcedata):
spec = {
'channels': [
{
'name': 'signal',
'samples': [
{
'name': 'signal',
'data': sourcedata['signal']['bindata']['sig'],
'modifiers': [
{
'name': 'mu',
'type': 'normfactor',
'data': None
}
]
},
{
'name': 'bkg1',
'data': sourcedata['signal']['bindata']['bkg1'],
'modifiers': [
{
'name': 'coupled_histosys',
'type': 'histosys',
'data': {'lo_data': sourcedata['signal']['bindata']['bkg1_dn'], 'hi_data': sourcedata['signal']['bindata']['bkg1_up']}
}
]
},
{
'name': 'bkg2',
'data': sourcedata['signal']['bindata']['bkg2'],
'modifiers': [
{
'name': 'coupled_histosys',
'type': 'histosys',
'data': {'lo_data': sourcedata['signal']['bindata']['bkg2_dn'], 'hi_data': sourcedata['signal']['bindata']['bkg2_up']}
}
]
}
]
},
{
'name': 'control',
'samples': [
{
'name': 'background',
'data': sourcedata['control']['bindata']['bkg1'],
'modifiers': [
{
'name': 'coupled_histosys',
'type': 'histosys',
'data': {'lo_data': sourcedata['control']['bindata']['bkg1_dn'], 'hi_data': sourcedata['control']['bindata']['bkg1_up']}
}
]
}
]
}
]
}
pdf = Model(spec)
data = []
for c in pdf.spec['channels']:
data += sourcedata[c['name']]['bindata']['data']
data = data + pdf.config.auxdata
return data, pdf
[4]:
validation_datadir = '../../validation/data'
[5]:
source = json.load(open(validation_datadir + '/2bin_2channel_coupledhisto.json'))
data, pdf = prep_data(source['channels'])
print(data)
init_pars = pdf.config.suggested_init()
par_bounds = pdf.config.suggested_bounds()
unconpars = pyhf.optimizer.unconstrained_bestfit(pyhf.utils.loglambdav, data, pdf, init_pars, par_bounds)
print('parameters post unconstrained fit: {}'.format(unconpars))
conpars = pyhf.optimizer.constrained_bestfit(pyhf.utils.loglambdav, 0.0, data, pdf, init_pars, par_bounds)
print('parameters post constrained fit: {}'.format(conpars))
pdf.expected_data(conpars)
INFO:pyhf.pdf:Validating spec against schema: /home/jovyan/pyhf/pyhf/data/spec.json
INFO:pyhf.pdf:adding modifier mu (1 new nuisance parameters)
INFO:pyhf.pdf:adding modifier coupled_histosys (1 new nuisance parameters)
[170.0, 220.0, 110.0, 105.0, 0.0]
parameters post unconstrained fit: [1.05563069e-12 4.00000334e+00]
parameters post constrained fit: [0. 4.00000146]
[5]:
array([ 1.25000007e+02, 1.60000022e+02, 2.10000022e+02, -8.00631284e-05,
4.00000146e+00])
[6]:
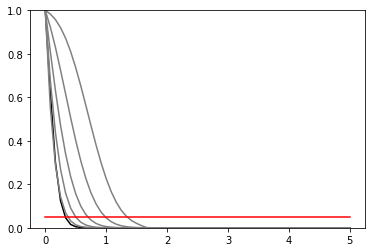
def plot_results(test_mus, cls_obs, cls_exp, poi_tests, test_size = 0.05):
plt.plot(poi_tests,cls_obs, c = 'k')
for i,c in zip(range(5),['grey','grey','grey','grey','grey']):
plt.plot(poi_tests, cls_exp[i], c = c)
plt.plot(poi_tests,[test_size]*len(test_mus), c = 'r')
plt.ylim(0,1)
def invert_interval(test_mus, cls_obs, cls_exp, test_size = 0.05):
point05cross = {'exp':[],'obs':None}
for cls_exp_sigma in cls_exp:
yvals = cls_exp_sigma
point05cross['exp'].append(np.interp(test_size,
list(reversed(yvals)),
list(reversed(test_mus))))
yvals = cls_obs
point05cross['obs'] = np.interp(test_size,
list(reversed(yvals)),
list(reversed(test_mus)))
return point05cross
poi_tests = np.linspace(0, 5, 61)
tests = [pyhf.utils.hypotest(poi_test, data, pdf, init_pars, par_bounds, return_expected_set=True)
for poi_test in poi_tests]
cls_obs = np.array([test[0] for test in tests]).flatten()
cls_exp = [np.array([test[1][i] for test in tests]).flatten() for i in range(5)]
print('\n')
plot_results(poi_tests, cls_obs, cls_exp, poi_tests)
invert_interval(poi_tests, cls_obs, cls_exp)
INFO:pyhf.pdf:Validating spec against schema: /home/jovyan/pyhf/pyhf/data/spec.json
INFO:pyhf.pdf:adding modifier mu (1 new nuisance parameters)
INFO:pyhf.pdf:adding modifier coupled_histosys (1 new nuisance parameters)
[170.0, 220.0, 110.0, 105.0, 0.0]
parameters post unconstrained fit: [1.05563069e-12 4.00000334e+00]
parameters post constrained fit: [0. 4.00000146]
[6]:
{'exp': [0.37566312243018246,
0.49824494455027707,
0.7023047842202288,
0.9869744452422563,
1.3443167343146392],
'obs': 0.329179494864517}